n6 is heading to FoG London and SLAS2026!
COME MEET US LIVE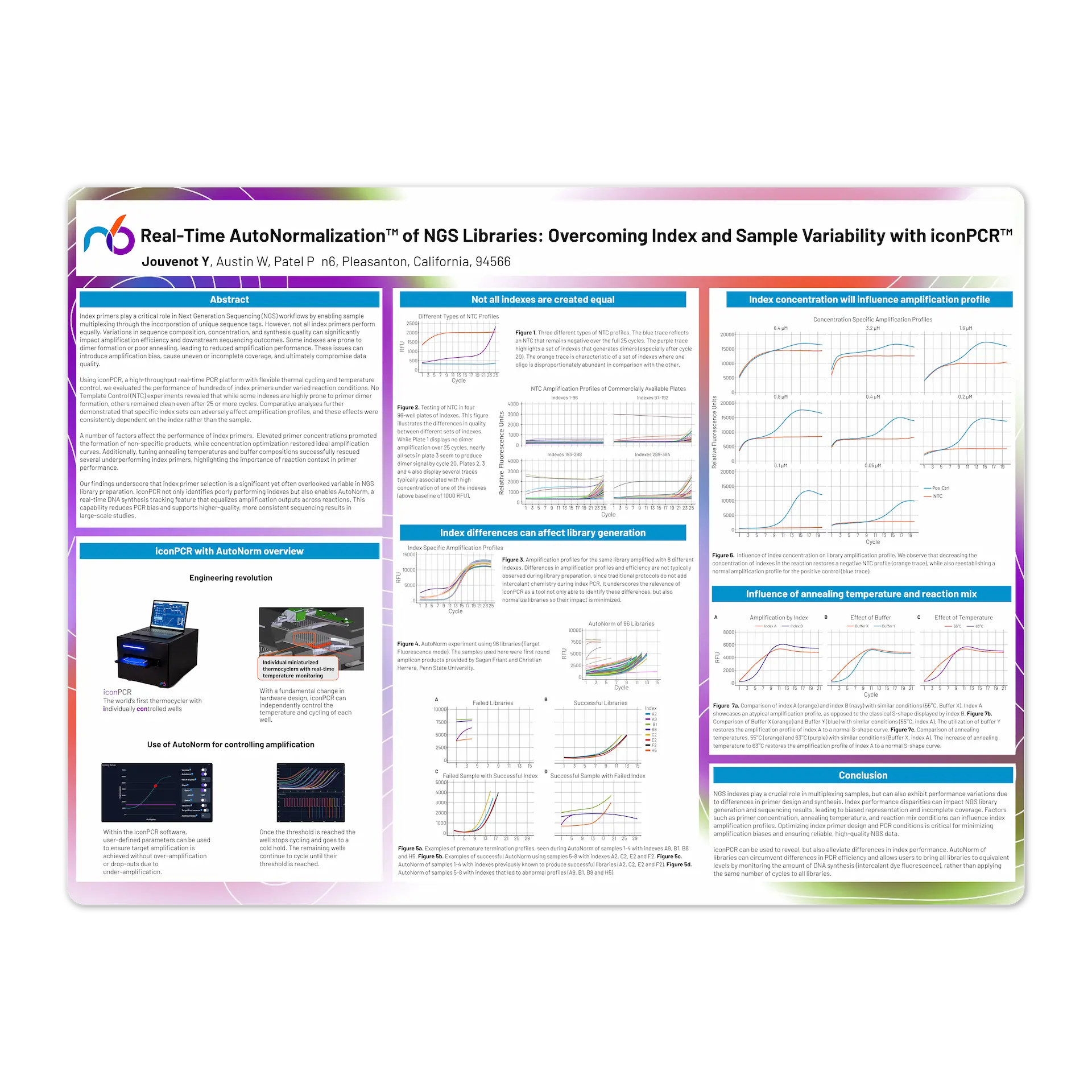
Index primers play a critical role in Next Generation Sequencing (NGS) workflows by enabling sample multiplexing through the incorporation of unique sequence tags. However, not all index primers perform equally. Variations in sequence composition, concentration, and synthesis quality can significantly impact amplification efficiency and downstream sequencing outcomes. Some indexes are prone to dimer formation or poor annealing, leading to reduced amplification performance. These issues can introduce amplification bias, cause uneven or incomplete coverage, and ultimately compromise data quality.
Using iconPCR, a high-throughput real-time PCR platform with flexible thermal cycling and temperature control, we evaluated the performance of hundreds of index primers under varied reaction conditions. No Template Control (NTC) experiments revealed that while some indexes are highly prone to primer dimer formation, others remained clean even after 25 or more cycles. Comparative analyses further demonstrated that specific index sets can adversely affect amplification profiles, and these effects were consistently dependent on the index rather than the sample.
A number of factors affect the performance of index primers. Elevated primer concentrations promoted the formation of non-specific products, while concentration optimization restored ideal amplification curves. Additionally, tuning annealing temperatures and buffer compositions successfully rescued several underperforming index primers, highlighting the importance of reaction context in primer performance.
Our findings underscore that index primer selection is a significant yet often overlooked variable in NGS library preparation. iconPCR not only identifies poorly performing indexes but also enables AutoNorm, a real-time DNA synthesis tracking feature that equalizes amplification outputs across reactions. This capability reduces PCR bias and supports higher-quality, more consistent sequencing results in large-scale studies.


