Upcoming webinar: How to optimize low-input cfDNA library prep for targeted sequencing sensitivity
REGISTER for MARCH 25Explore what’s possible with n6: from RNA-seq to metagenomics, whole genomes, single cells, and even the wildest “other” experiments, discover application freedom without workflow headaches. iconPCR with AutoNorm sets a new standard for NGS prep—seamlessly combining quantification, normalization, and amplification for every individual sample, whatever the input, quality, or project complexity. Whether your challenge is low input, high variability, or just plain speed, you’ll find every application gets the same balanced, high-quality libraries and dramatically streamlined hands-on time. One platform, endless scientific stories—now let’s find out where iconPCR takes your research next.
talk to an application specialist
Unlock the full story in your microbiome research with iconPCR™ and AutoNorm™—delivering next-level 16S sequencing for every sample, every run. Skip the biases and dropped species of traditional PCR: thanks to real-time, per-well control, you’ll capture truer community profiles, reduce chimeras by up to 4x, and reveal more unique amplicon sequence variants (ASVs)—including the rare and elusive. Whether you’re running short or full-length 16S, you get balanced, deeply resolved libraries and hands-off pooling—no matter the input or sample source. Discover how easy it is to leave artifacts and normalization headaches behind, so your data reflects the real diversity in every tube.
learn more
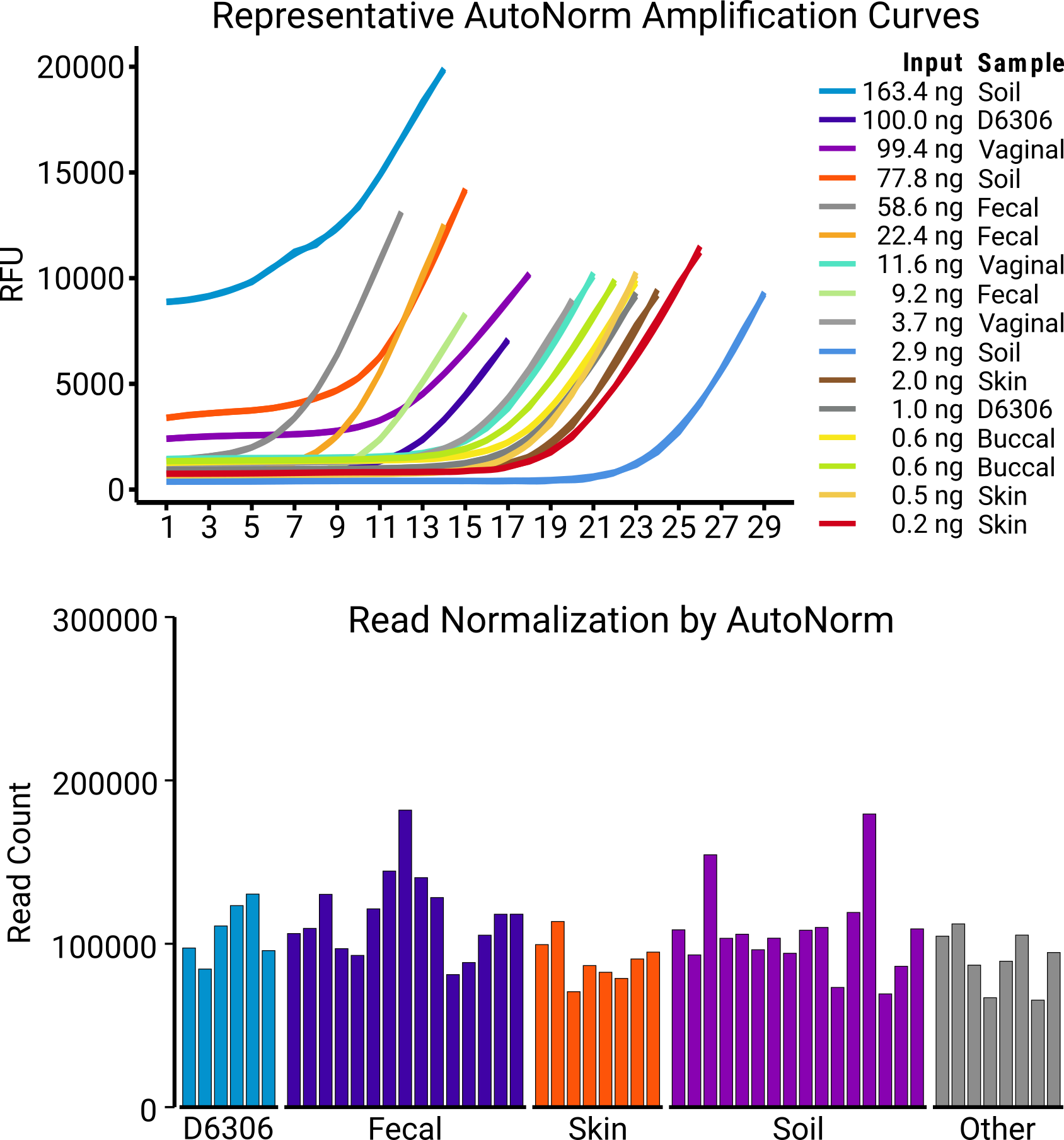
Curious what happens when you throw the whole ecosystem at iconPCR? Check out our “kitchen sink” experiment, where we threw every matrix (kitchen sink swab coming soon, maybe) in a single 96-well run—skin, soil, fecal, mouth, controls—each one normalized, amplified, and balanced perfectly, no extra hands-on required. The result: beautifully resolved, well-balanced reads from the most complex mixtures science can throw at you.
learN moreRescue Every Read: FFPE & Degraded Sample Success, Stress-Free
The phrase “challenging sample” just got taken off your worry list. iconPCR with AutoNorm transforms FFPE (Formalin-Fixed, Paraffin-Embedded) and other degraded specimens from data risk to NGS goldmine. Where traditional protocols can over- or under-cycle, leading to dropouts, high duplication rates, or ghost reads, iconPCR uses real-time fluorescence to ensure every library gets just the right push—no matter DIN or input variability. Rescue poor-quality prep with high-confidence amplification, optimal yield, and balanced normalization into your capture or sequencing pool. Run your ultra-precious, low-yield samples plus the “easy” ones side-by-side in one smart, streamlined workflow—with reliable results every time and no extra cost on hands or reagents.


Unlocking True Expression:
RNA-Seq Without the Guesswork
Say goodbye to overamplification and wavering data quality—iconPCR with AutoNorm brings harmony to RNA-seq library prep. Instead of spending precious time quantifying inputs and splitting samples based on quality, every well is individually optimized with real-time feedback, stopping amplification exactly where it should. That means no more under- or over-cycling, even for those tricky FFPE or degraded samples. The result? More genes detected, lower PCR artifacts and duplication rates, boosted data quality, and an effortless workflow that lets you process mixed quality samples all at once—while reducing hands-on time by up to 50%. Why settle for mediocre melodies when you can have a symphony of high-fidelity RNA-seq data?
LEARN MORE ABOUT RNA-SEQUENCINGEvery Cell Counts: Single Cell RNA-Seq, Perfected
Single cell experiments demand precision, speed, and the confidence that each cell's expression truly shines. iconPCR with AutoNorm takes the stress out of single cell RNA-seq, eliminating the need to know your cell count or RNA content up front. Our platform stops each amplification reaction at the perfect point using real-time fluorescence, so you can run all your single cell libraries—regardless of input or complexity—side by side in one instrument. Fewer runs, fewer errors, zero loss of data quality, and all the flexibility you crave. Fire up your single cell explorations knowing iconPCR has your back (and your UMAP clusters).
LEARN MORE ABOUT RNA-SEQUENCING

Whole Genome, Whole Control:
WGS Simplified
Whole genome sequencing (WGS) projects, meet your new timesaver. iconPCR with AutoNorm ensures every sample reaches optimal amplification—no matter the starting DNA quantity or integrity. Ditch multiple PCR runs and endless quantification for a one-step process, where input variation is no longer a limitation but a reason to run everything together. With automated normalization in every well, you get balanced libraries, consistent yields, and avoid the mysteries of over- or under-amplification, especially for challenging samples like FFPE. Result: better coverage, less waste, and sequencing pools so balanced you’ll want to show them off.
REAd the app noteOpen the Possibilities: From ChIP-Seq to Custom Assays
Why box yourself in? iconPCR™ was built for versatility. Whether you’re tackling ChIP-seq, Cut&Tag, spatial transcriptomics, forensic science—or innovating your own assay—our platform’s flexible temperature and protocol control, combined with per-well real-time normalization, lets you mix, match, and develop new workflows quickly. Imagine running up to 96 different experimental conditions at once or standardizing tricky low-input or unusual sample types. Need to compare index performance or balance high-plex pools? It’s all at your fingertips.More applications and data coming soon but reach out if you want to talk emerging and in-development applications!Same button as top of page “talk to an application scientist”

How well do you know your indexes? Utilize iconPCR for index PCR to reveal differences in performance and help alleviate issues.
Indexes are not created equal nor do they perform the same in all conditions. Some designs will be prone to dimerisation, some will be at different concentrations, some will not be optimized at the right temperature. Differences in index performance may affect NGS libraries and sequencing results.
READ THE TECH NOTEDesign, Optimize, Repeat-Without the Wait
iconPCR empowers assay development with true Design of Experiments (DOE) flexibility—96 wells, 96 independent PCR conditions, on one plate. Eliminate split-plot headaches, test gradients and reagent tweaks side-by-side, and slice hands-on time by up to 80%. AutoNorm makes every well count, delivering real data for smarter assay iteration and rapid optimization. Get your answers (and your weekends) back, while saving time and consumables—all in a single, streamlined run.
READ THE TECH NOTE