n6 is heading to FoG London and SLAS2026!
COME MEET US LIVE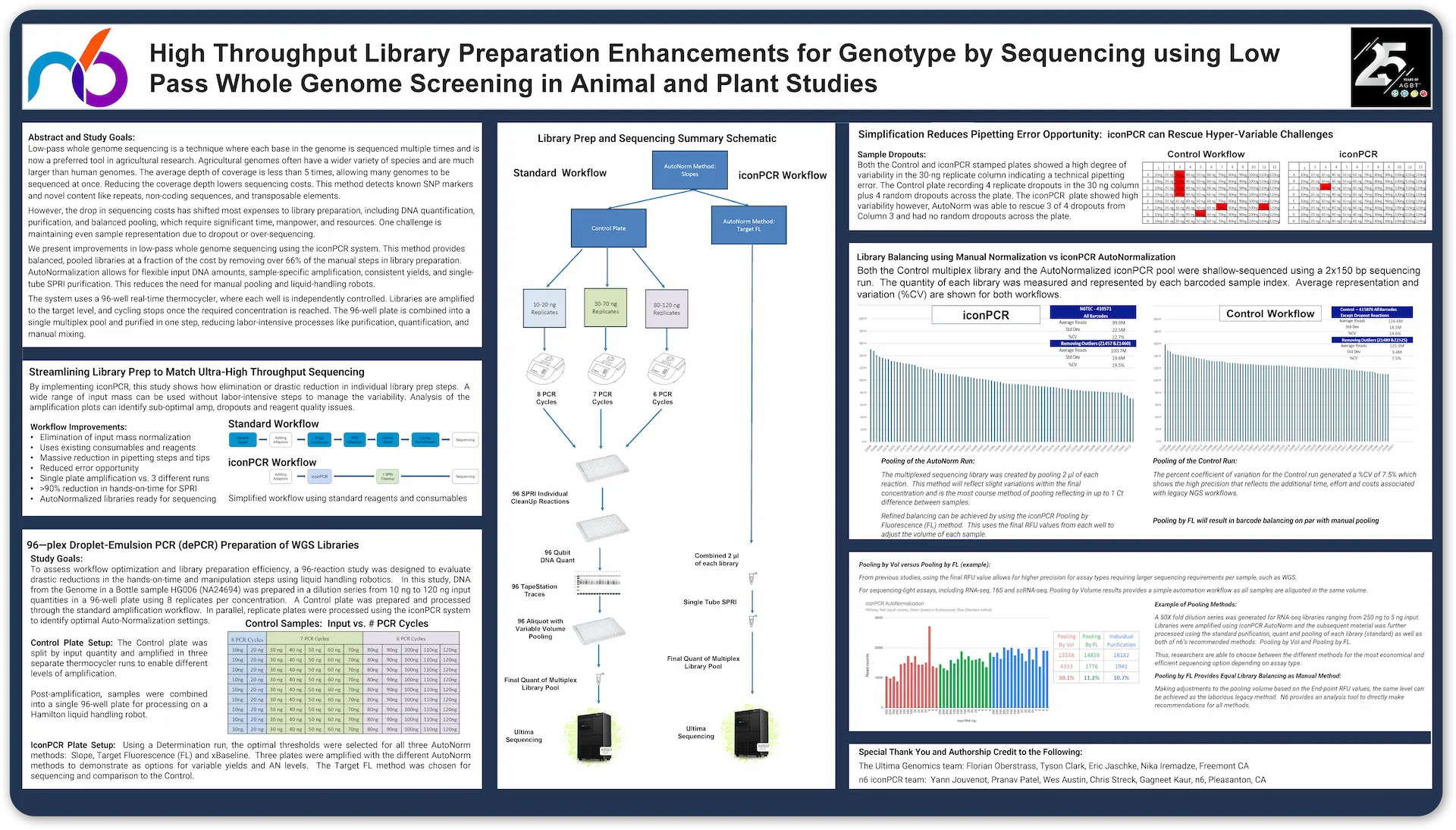
Low-pass whole genome sequencing is a technique where each base in the genome is sequenced multiple times and is now a preferred tool in agricultural research. Agricultural genomes often have a wider variety of species and are much larger than human genomes. The average depth of coverage is less than 5 times, allowing many genomes to be sequenced at once. Reducing the coverage depth lowers sequencing costs. This method detects known SNP markers and novel content like repeats, non-coding sequences, and transposable elements.
However, the drop in sequencing costs has shifted most expenses to library preparation, including DNA quantification, purification, and balanced pooling, which require significant time, manpower, and resources. One challenge is maintaining even sample representation due to dropout or over-sequencing.
We present improvements in low-pass whole genome sequencing using the iconPCR system. This method provides balanced, pooled libraries at a fraction of the cost by removing over 66% of the manual steps in library preparation. AutoNormalization allows for flexible input DNA amounts, sample-specific amplification, consistent yields, and singletube SPRI purification. This reduces the need for manual pooling and liquid-handling robots.
The system uses a 96-well real-time thermocycler, where each well is independently controlled. Libraries are amplified to the target level, and cycling stops once the required concentration is reached. The 96-well plate is combined into a single multiplex pool and purified in one step, reducing labor-intensive processes like purification, quantification, and manual mixing.


