Modern NGS runs are expensive propositions. A single lane on a NovaSeq X 25B flow cell can cost $15,000-$20,000, while high-throughput runs routinely reach $48,000 for 10 billion reads. When you factor in sample preparation costs averaging $50-$200 per sample, library preparation reagents, labor, and bioinformatics analysis, the total investment quickly escalates.
But here's what makes this particularly painful: most sequencing failures originate during library preparation, not sequencing itself. Research shows that 80% of mutation detection failures in clinical samples can be attributed to pre-PCR processing errors. The culprits are well-known but poorly controlled: overamplification and underamplification during PCR.
Where Traditional PCR Goes Wrong
Legacy thermocyclers force laboratories into a fundamentally flawed approach: guessing the right number of PCR cycles. This constraint has plagued NGS workflows since their inception, leading to predictable but costly consequences:
Overamplification creates PCR duplicates, chimeric sequences, and artifacts that consume expensive sequencing reads without providing useful data. When libraries are over-cycled, primer depletion causes PCR products to anneal to each other, forming high molecular weight artifacts that appear as "PCR bubbles" on electropherograms. These artifacts can consume 50-60% of sequencing reads, effectively wasting half of an expensive sequencing run.
Underamplification results in insufficient library yield, leading to dropouts, failed QC, and sample re-queues. This is particularly problematic for challenging samples like FFPE tissues, where degradation rates can reach 55% and success rates are notoriously unpredictable.
Research demonstrates that PCR amplification introduces multiple sources of error and bias that distort sequencing results. GC bias, stochastic amplification effects, template switching, and polymerase errors all compound during traditional fixed-cycle PCR, ultimately degrading the quality of expensive sequencing data.
The Brute Force Status Quo is Failing at Scale
The genomics industry is scaling exponentially—from thousands of samples to billions. Population genomics initiatives like the UK Biobank (500K genomes) and the NIH All of Us program (1M participants) represent just the beginning. Yet the tools powering this expansion are fundamentally unchanged from two decades ago.
Legacy thermocyclers weren't designed for this complexity or scale. They provide uniform heating across all wells, no real-time feedback, and no ability to optimize individual samples. This forces laboratories into inefficient batching workflows, where sample variability within a single plate leads to compromised results.
The consequences are becoming more severe as sequencing moves into clinical applications, where failed libraries mean delayed patient care, repeated biopsies, and potentially missed therapeutic windows. In research contexts, failed libraries mean missed publication deadlines, wasted grant funds, and unreproducible results.
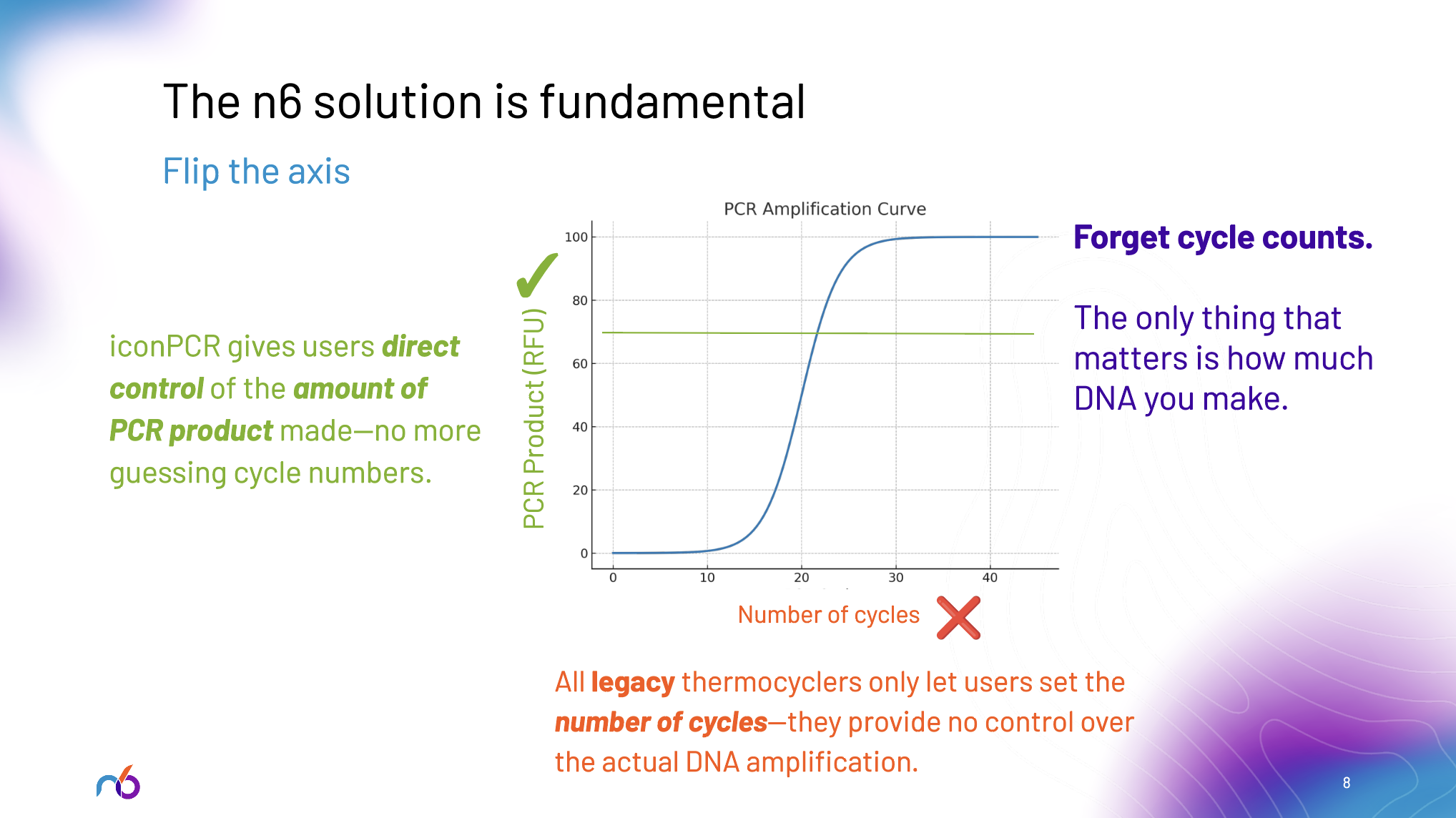
n6's Fundamental Solution: Flipping the Axis
n6 has solved this problem by fundamentally changing how PCR amplification works. Instead of constraining laboratories to fixed cycle numbers, iconPCR™ and AutoNorm™ flip the axis entirely: choose the fluorescence target that corresponds to your desired DNA yield, and let the instrument determine the optimal number of cycles for each individual sample.
This isn't an incremental improvement—it's a complete reimagining of how amplification should work in the high throughput genomics age. iconPCR provides 96 independently controlled thermal zones with real-time fluorescence monitoring in each well. AutoNorm dynamically adjusts cycle numbers per well to normalize output concentrations across all samples, eliminating the guesswork that leads to expensive failures.
The result: amplification, quantification, and normalization happen simultaneously in a single automated step. No more pre-PCR quantification. No more post-PCR normalization kits. No more individual SPRI cleanups—just one cleanup on the final pool instead of 96 separate cleanups.
Proven Results from Leading Institutions
Major genomics centers are already seeing the impact. Dr. Anja Mezger at SciLifeLab reports that "iconPCR will help us streamline processes by eliminating QC and normalization steps, while also improving data quality by preventing over-amplification of samples". At UC San Diego's IGM Genomics Center, Dr. Kristen Jepsen notes that "by more accurately balancing libraries, we can increase throughput and reduce false negatives and positives in our metagenomic studies".

The data back up these testimonials. iconPCR demonstrates reduced PCR duplicates, fewer chimeric sequences, and improved rescue of challenging samples including FFPE and low-input materials. For RNA-seq applications, users report higher gene counts, lower duplication rates, and better preservation of biological data.
The Economic Reality: Prevention vs. Waste
The math is stark. iconPCR saves over $1,000 per 96-sample run before samples even reach the sequencer. For a medium-throughput lab running eight 96-sample batches per month, monthly savings exceed $84,000, translating to over $1M annually. These savings come from eliminating reagent waste, reducing hands-on time, and preventing the expensive re-runs that plague traditional workflows.
But the real value isn't just in cost reduction—it's in risk mitigation. Every prevented failure saves the cost of a car. Every successful library on the first attempt means faster turnaround times, better resource utilization, and more reliable results.
How Much Longer Can the Industry Wait?
The question facing every NGS laboratory is simple: How can you afford not to fix this problem? How can you afford compromised data quality when algorithms and AI systems depend on clean inputs? How can you afford inefficient workflows when genomics is moving from research curiosity to clinical necessity? How can you afford the uncertainty of re-runs and failed libraries when patients and discovery timelines depend on reliable results?
PCR amplification has been genomics' enemy for two decades. The artifacts, bias, and waste it introduces have been tolerated because there was no alternative. But that era is ending.
iconPCR represents what the genomics industry has needed but never had: intelligent, per-sample control that eliminates the fundamental causes of library prep failure. It's not just a better thermocycler—it's an engineering solution to a problem the industry had deemed impossible to solve.
The future of genomics will be defined by tools that respect every read, every sample, and every dollar invested in generating data. Labs that continue using legacy approaches are essentially choosing to waste the cost of a car on every failed run.
The technology exists. The validation is complete. The question is: how much longer can your lab afford to wait?
For an industry racing toward population-scale genomics and precision medicine, the answer should be clear. The cost of inaction—in dollars, time, and missed opportunities—is one that labs should no longer accept to bear anymore.




